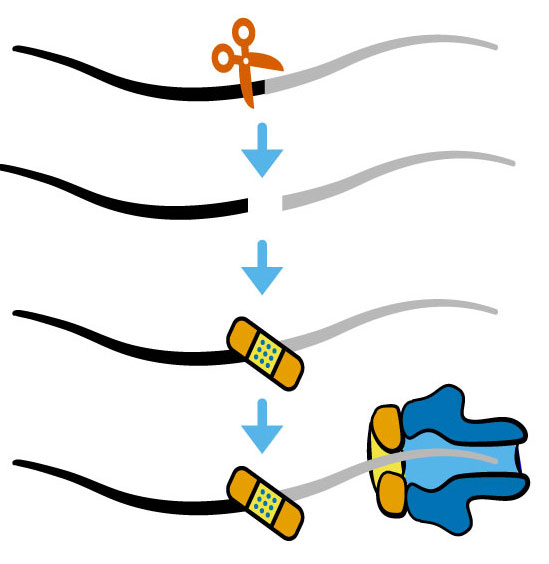
RNA damage and repair
Like DNA repair systems that surveil and repair genome damage to preserve the genetic code, the products of RNA damage are substrates for coupled end modification and processing steps. But unlike our deep understanding of the DNA damage response, we are only beginning to learn how RNA damage, end modification, and processing are integrated in the RNA damage response to orchestrate RNA processing for regulatory control.
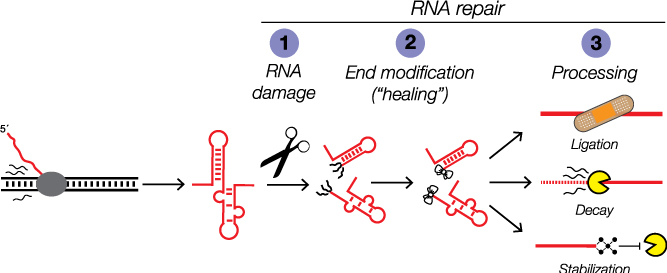
Growing evidence indicates RNA repair activities can be linked together to create elegant forms of post-transcriptional control. We and others discovered examples of RNA processing by “kinase-mediated decay” wherein RNAs with 5´-OH termini are degraded by sequential 5´-OH phosphorylation and 5´-3´ degradation by a 5´-phosphate-dependent exoribonuclease. Given the diversity of RNA repair activities, other dependencies between RNA end modification and processing could be used for new forms of post-transcriptional regulation.
Our research program tackles open questions in RNA repair by combining genetics, biochemistry, and bioinformatics to explore the causes and consequences of RNA damage. In doing so, we have uncovered new concepts in post-transcriptional regulation and are uniquely positioned to continue to explore the surprising roles of the RNA damage response in biology.
RNA sequencing approaches to study RNA damage & repair
We previously developed methods to capture the 2’,3’-cyclic phosphate and 5’-OH products of RNA decay and used them to characterize products of spontaneous and enzyme-catalyzed RNA cleavage.

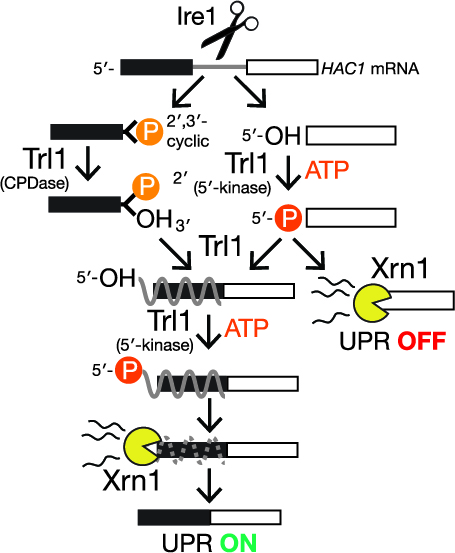
Recently we developed an approach leveraging nanopore sequencing to identify RNA 2´-phosphorylation — a bona fide biochemical mark of RNA repair. This approach enabled unambiguous detection of repair events on tRNA products of ligation and the Hac1 mRNA, which is ligated during the unfolded protein responses. We also identified a collection of mRNAs that generate similar signals during nanopore sequencing, suggesting they are also targets of repair. We continue to apply this approach to characterize RNA repair events during stress conditions.
tRNA splicing
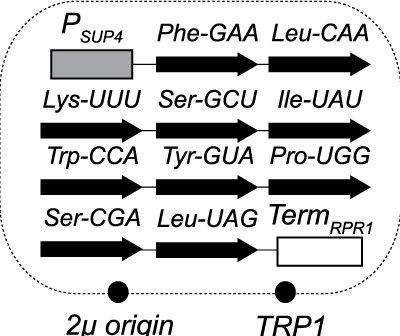
We recently generated budding yeast cells that lack normally essential compo-nents of RNA repair. RNA ligase enzymes catalyze rejoining of RNA after cleavage of phosphodiester linkag-es. RNA ligation in budding yeast is catalyzed by two separate enzymes that ligate exons of tRNA during splicing and HAC1 mRNA during activation of the unfolded protein response. The yeast RNA ligase Trl1 joins 2’,3’-cyclic phosphate and 5’-OH RNA fragments 32, yielding a phosphodiester linkage with a 2’-phosphate that is subsequently removed by the 2’-phosphotransferase Tpt1. Inspired by previous work in C. elegans, we rescued the lethality of TRL1 and TPT1 deletions in budding yeast by expressing intron-less versions of the 10 normally intron-containing tRNAs, indicating this repair pathway does not have additional essential functions. These cells are critical reagents that have opened the door to new studies of eukaryotic RNA repair. We are currently expanding this approach to study the breadth of RNA repair across evolution.
It is noteworthy that this approach was unable to bypass the essentialty of the TSEN tRNA splicing complex, resonating with earlier work from the Hopper lab. We are using the genetic bypass strategy to identify other essential functions of TSEN, which may shed light on its role in the human neurodegenerative disease Pontocerebellar hypoplasia.
Unfolded protein response
We recently discovered a previously unappreciated layer of post-transcriptional regulation during the unfolded protein response (UPR). In the UPR, endoplasmic reticulum stress activates a large transcriptional program to increase folding capacity. In the budding yeast UPR, Ire1 excises an intron from the HAC1 mRNA and the exon products of cleavage are ligated by Trl1, yielding an mRNA encoding a transcription factor whose translation induces hundreds of stress-response genes. Moreover, robust base-pairing between the 5’-exon and intron inhibits HAC1 translation Using cells with mutations in RNA repair and decay enzymes, we showed that kinase-mediated decay of HAC1 splicing intermedi-ates both suppresses and activates the UPR. Together with recent studies our findings support a new paradigm in cell signaling: analogous to kinase-mediated signal transduction through protein substrates, the UPR is controlled by kinase-mediated signal transduction through RNA substrates. These studies provide a strong conceptual foundation for understanding new couplings between RNA damage and repair.