Hesselberth Lab

We are a research laboratory of RNA biologists, technology developers, and data analysts focused on discovering and translating fundamental principles of RNA regulation.
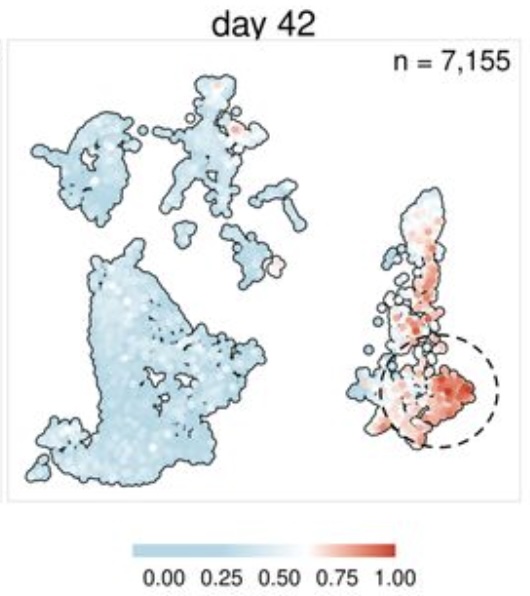
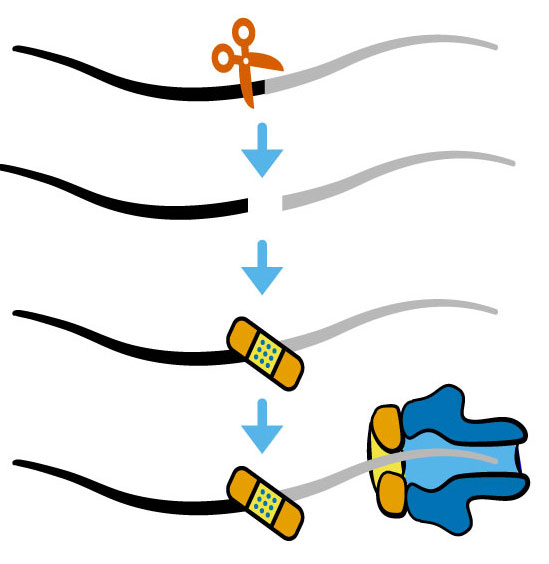
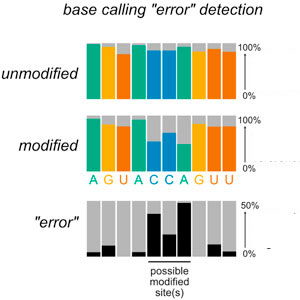
A major effort in the lab is to understand how RNA damage and repair are integrated with stress responses by combining method development, bioinformatics, genetics, biochemistry, and cell biology. We developed new sequencing approaches to identify damaged RNAs and new model systems to understand the causes and consequences of RNA damage. Our studies have led to new concepts in post-transcriptional regulation with broad implications for understanding the role of RNA damage and repair.
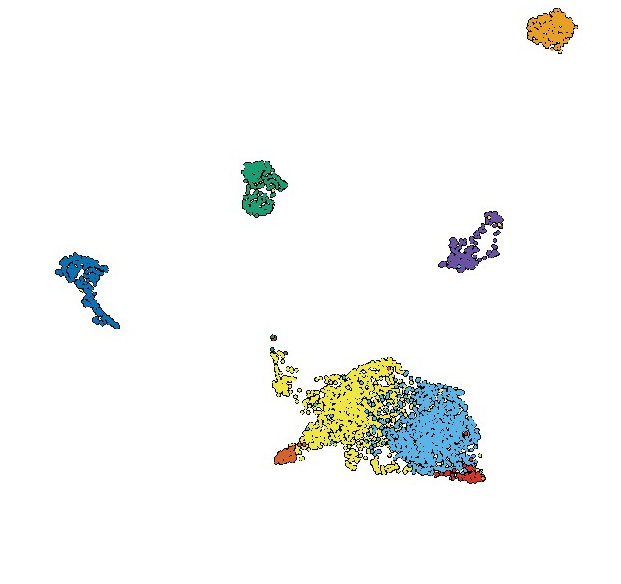
A second focus is on molecular tools that enhance single-cell and spatial transcriptomic experiments. Our recent advances include the measurement of enzyme activity and inhibition in single-cell assays and tracking protein molecules and measuring associated gene expression responses in animal tissue.
Our newest efforts focus on RNA therapeutics and vaccines. One is a collaboration with clinicians at Children’s Hospital Colorado to develop RNA therapies for pediatric neurological diseases, and a second emphasis is on development of mRNA vaccines that promote unique immune responses.
See the projects link above for more detail on these studies.
selected publications
news
| Jul 2, 2024 |
|
|---|---|
| Jul 9, 2023 |
|
| Dec 1, 2022 |
|