Molecular technology development
Single-cell biochemistry: a new approach to measure functional heterogeneity
New methods to study heterogeneity at cellular resolution in complex tissues are transforming our understand-ing of human biology and disease. These approaches measure differences in gene expression, chromatin ac-cessibility, and protein levels across thousands to millions of cells to understand developmental trajectories of tissues, tumors, and whole organisms. But these methods rely on measurements of static levels of DNA, RNA, and proteins, and fail to capture dynamic biochemical activities that underlie complex cellular functions. Instead of developing more direct readouts of cellular function, the field has focused on inferring functional status from measurement of mRNA abundance and chromatin accessibility in single cells.
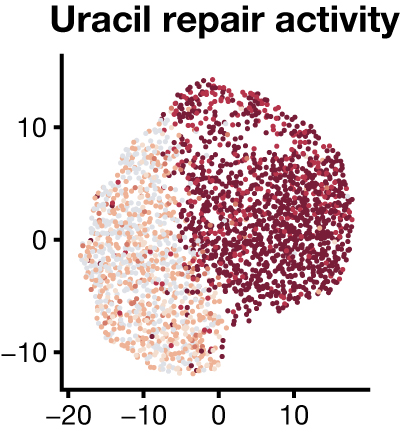
To accelerate the study of biochemical heterogeneity among single cells, we are developing functional assays as a new modality for single-cell experiments. Instead of measuring the abundance of molecules – levels of DNA, RNA, or protein – from single cells and predicting cell functional states, our key innovation is to directly measure enzymatic activities in single cells by measuring the conversion of substrates to products by single cell extracts in a high-throughput DNA sequencing experiment. Our approach is compatible with existing platforms that measure gene expression in thousands to millions of individual cells and enables many different enzymatic activities to be measured simultaneously.
Charting tissue dissmination with molecular tracking devices
The dissemination of molecules through tissue establishes signaling gradients that drive complex developmental patterns, enables rapid responses to infection, and dictates the distribution of therapeutic drugs. However, our understanding of this molecular cartography would be vastly improved by charting molecular traffic throughout organ systems and cataloguing cell and tissue responses upon arrival and sensing.
We recently developed “molecular tracking devices” – protein-DNA conjugates that are stable during tissue transit in vivo – to follow the distribution, acquisition, and retention of antigen in the immune system during an immune response. Molecular tracking devices enable previously inaccessible measurements of gene expression responses to signaling gradients and ligand-receptor interactions in living animals. We are expanding this approach to study the dynamics and concentrations of tracking devices within tissue and associated gene expression responses caused by receptor activation.
Funding
- CU ASPIRE (with Beth Tamburini, Tem Morrison, and Jenna Guthmiller)
- NIH/NIA Transformative R01
- NIH/NIAID R21